How Owkin Diagnostics tackles the challenge of generalizability
In the dynamic landscape of AI diagnostics, one crucial aspect that is often overlooked is generalizability – the reliability and consistency of an algorithm across various settings.
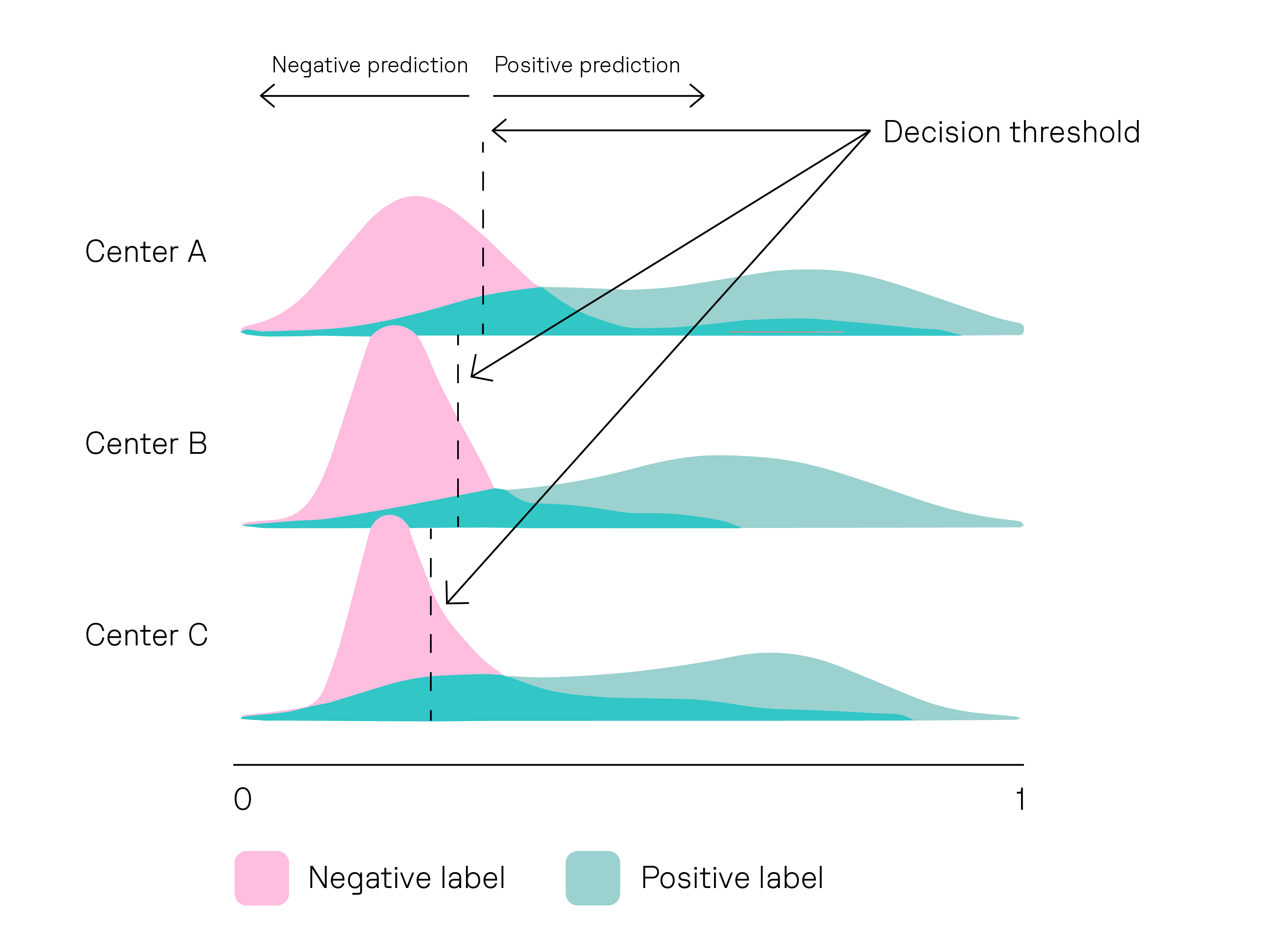
Settings can differ in different ways. For example, different laboratories might use different scanner brands to capture images of the tissue samples. Or staining techniques might differ between centers1.
To show high generalizability, an AI diagnostic (like Owkin’s MSIntuit CRC) would need to reliably detect a biomarker of interest, regardless of the setting in which it was deployed.
Ensuring generalizability across laboratory settings is essential for the widespread adoption of AI diagnostics in clinical practice. It allows healthcare providers to confidently use a diagnostic in various laboratory settings, knowing that it will consistently deliver accurate and reliable results.
Owkin's access to rich datasets, combined with our expertise in AI and machine learning, is fueling remarkable advancements in the performance and robustness of our models1,2,3. For example, we have recently developed Phikon, a cutting-edge histology foundation model.
Foundation models like Phikon provide exciting possible ways to enhance generalizability, but existing AI models are still insufficient to fully bridge the gap, leaving this issue an ongoing research question within the field.

How can we adapt our models to ensure sufficient performance quality in an unknown setup?
Owkin’s team is leading the field in this generalizability challenge. This month, dozens of machine learning engineers, data scientists, and biostatisticians collaborated on a two-day hackathon on generalizability.
We concentrated on how to adapt our models for practical use in a real-world setting (for example through model calibration). Our findings are exciting:
- We found we could potentially reduce the size of a required calibration set by using richer information and prior knowledge, beyond model output such as the training dataset.
- Out-of-domain generalization techniques(a way of training models to work on previously unseen domains of data - compared to their training data) can even work on unlabelled data (data that has not been annotated by humans), but they tend to not work as well as methods using labeled data.
- True generalizability can only be achieved by paying close attention to all sources of variability during both the training stage and when being used in the real world (inference stage).
Lionel Guillou, VP Technology development & Data - Diagnostics, Owkin, said that:
At Owkin, we confront the challenge of generalizability head-on, recognizing its pivotal role in fortifying the reliability and utility of our AI tools. Proposing innovative solutions to this challenge will be key in building and maintaining trust with clinicians that use our AI tools.
Conclusion
In our ongoing quest for improvement, we stay dedicated to enhancing generalizability, recognizing its crucial role in AI-based diagnostics. As we navigate the intricate landscape of AI-driven healthcare, we're prepared to overcome obstacles and pioneer advancements in diagnostic accuracy and reliability.
1 Saillard, C., Dubois, R., Tchita, O. et al. Validation of MSIntuit as an AI-based pre-screening tool for MSI detection from colorectal cancer histology slides. Nat Commun 14, 6695 (2023). https://doi.org/10.1038/s41467-023-42453-6
2 Imran, A., Scaling Self Supervised Learning for Histology: introducing Phikon. Hugging Face (2023). https://huggingface.co/blog/EazyAl/phikon
3 Schiratti, J-B., Di Proietto, V., Andrier, E., Mantiero, D. UBC Ocean Kaggle competition – Meet the winners. Owkin (2024). https://www.owkin.com/blogs-case-studies/ubc-ocean-kaggle-competition-meet-the-winners
4 Jindong, W. et al. Generalizing to Unseen Domains: A Survey on Domain Generalization. arXiv (2022). https://arxiv.org/pdf/2103.03097.pdf
5 Mostafa, J.,Domain Generalization in Computational Pathology: Survey and Guidelines. arXiv (2023). https://arxiv.org/abs/2310.19656
6 Roschewitz, M., Khara, G., Yearsley, J. et al. Automatic correction of performance drift under acquisition shift in medical image classification. Nat Commun 14, 6608 (2023). https://doi.org/10.1038/s41467-023-42396-y





